Command line tutorial¶
This tutorial will walk you through the usage of the lib5c
command-line tools.
Follow along in Google colab¶
You can run and modify the cells in this notebook tutorial live using Google colaboratory by clicking the link below:
To simply have all the cells run automatically, click
Runtime > Run all in the colab toolbar.
Make sure lib5c is installed¶
Inside a fresh virtual environment, run
In [1]:
!pip install lib5c
Requirement already satisfied: lib5c in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (0.5.3)
Requirement already satisfied: numpy>=1.10.4 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.14.2)
Requirement already satisfied: pandas>=0.18.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.22.0)
Requirement already satisfied: statsmodels>=0.6.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.8.0)
Requirement already satisfied: dill>=0.2.5 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.2.7.1)
Requirement already satisfied: luigi>=2.1.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.7.3)
Requirement already satisfied: matplotlib>=1.4.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.2.2)
Requirement already satisfied: python-daemon<2.2.0,>=2.1.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.1.2)
Requirement already satisfied: powerlaw>=1.4.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.4.3)
Requirement already satisfied: interlap>=0.2.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.2.6)
Requirement already satisfied: seaborn>=0.8.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.8.1)
Requirement already satisfied: decorator>=4.0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (4.2.1)
Requirement already satisfied: scikit-learn>=0.17.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.19.1)
Requirement already satisfied: scipy>=0.16.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.0.0)
Requirement already satisfied: python-dateutil in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from pandas>=0.18.0->lib5c) (2.7.2)
Requirement already satisfied: pytz>=2011k in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from pandas>=0.18.0->lib5c) (2018.3)
Requirement already satisfied: patsy in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from statsmodels>=0.6.1->lib5c) (0.5.0)
Requirement already satisfied: tornado<5,>=4.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from luigi>=2.1.1->lib5c) (4.5.3)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (2.2.0)
Requirement already satisfied: backports.functools-lru-cache in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.5)
Requirement already satisfied: subprocess32 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (3.5.3)
Requirement already satisfied: six>=1.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.11.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.0.1)
Requirement already satisfied: cycler>=0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (0.10.0)
Requirement already satisfied: setuptools in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (40.4.3)
Requirement already satisfied: docutils in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (0.14)
Requirement already satisfied: lockfile>=0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (0.12.2)
Requirement already satisfied: backports-abc>=0.4 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (0.5)
Requirement already satisfied: certifi in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (2018.1.18)
Requirement already satisfied: singledispatch in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (3.4.0.3)
Make a directory and get data¶
If you haven’t completed the pipeline tutorial yet, make a directory for the tutorial:
$ mkdir lib5c-tutorial
$ cd lib5c-tutorial
and prepare the example data in lib5c-tutorial/input as shown in the
pipeline tutorial.
In [2]:
!mkdir input
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE68582&format=file&file=GSE68582%5FBED%5FES%2DNPC%2DiPS%2DLOCI%5Fmm9%2Ebed%2Egz' | gunzip -c > input/BED_ES-NPC-iPS-LOCI_mm9.bed
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974095&format=file&file=GSM1974095%5Fv65%5FRep1%2Ecounts%2Etxt%2Egz' | gunzip -c > input/v65_Rep1.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974096&format=file&file=GSM1974096%5Fv65%5FRep2%2Ecounts%2Etxt%2Egz' | gunzip -c > input/v65_Rep2.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974099&format=file&file=GSM1974099%5FpNPC%5FRep1%2Ecounts%2Etxt%2Egz' | gunzip -c > input/pNPC_Rep1.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974100&format=file&file=GSM1974100%5FpNPC%5FRep2%2Ecounts%2Etxt%2Egz' | gunzip -c > input/pNPC_Rep2.counts
!sed -i '/Sox2\|Klf4/!d' input/BED_ES-NPC-iPS-LOCI_mm9.bed
mkdir: cannot create directory ‘input’: File exists
Note for Docker image users¶
If you are using lib5c from the Docker image, run:
$ docker run -it -v <full path to lib5c-tutorial>:/lib5c-tutorial creminslab/lib5c:latest
root@<container_id>:/# cd /lib5c-tutorial
and continue running all tutorial commands in this shell.
Trimming low-quality primers¶
We will first trim away low-quality primers by running
In [3]:
!lib5c trim -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -t trimmed/%s.counts trimmed/primers_trimmed.bed 'input/*.counts'
creating directory trimmed
Normalization¶
For this tutorial, we will first use Knight-Ruiz matrix balancing to correct our data for primer effects by running
In [4]:
!lib5c kr trimmed/pNPC_Rep2.counts kr/pNPC_Rep2.counts
loading counts
kr balancing
writing counts
creating directory kr
lib5c kr specifies that the specific subcommand we want is kr,
the subcommand for Knight-Ruiz balancing. To see all the available
subcommands, you can run
In [5]:
!lib5c -h
sub-commands
pipeline run entire pipeline
hic-extract extract chunks from Hi-C data
trim primer and countsfile trimming
outliers remove high spatial outliers
remove remove low-count primer-primer pairs
qnorm quantile normalization
express express normalization
kr knight-ruiz matrix balancing normalization
spline spline normalization
iced iced balancing normalization
determine-bins create binfiles from primerfiles
bin bin fragment-level counts
smooth smooth counts
expected compute expected models
variance estimate variances from obs values and exp models
pvalues call pvalues for interactions
threshold threshold p-values
qvalues perform multiple testing correction
interaction-score convert p-values to interaction scores
subtract subtract countsfiles
divide divide countsfiles
log log countsfiles
colorscale compute regional colorscales for comparing replicates
plot plot things
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
To see detailed help for the kr subcommand, you can run
In [6]:
!lib5c kr -h
usage: lib5c kr [-h] [-p PRIMERFILE] [-H] [-B] [-i MAX_ITERATIONS]
[-s IMPUTATION_SIZE]
infile outfile
positional arguments:
infile Input countsfile.
outfile Output countsfile.
optional arguments:
-h, --help show this help message and exit
-p PRIMERFILE, --primerfile PRIMERFILE
Primer file or bin file to use. If this flag is not
present, a .bed file will be searched for based on the
locations of the input files.
-H, --hang If multiple input files are specified, this flag will
cause the terminal to hang until all input files have
finished processing.
-B, --output_bias If this flag is present, the bias vectors will be
written to .bias files located next to the output
.counts files.
-i MAX_ITERATIONS, --max_iterations MAX_ITERATIONS
Maximum number of iterations. The default is 3000.
-s IMPUTATION_SIZE, --imputation_size IMPUTATION_SIZE
Size of window, in units of matrix indices, to use to
impute nan values in the original counts matrix. Pass
0 to skip imputation, which is the default behavior.
The two required positional command line arguments for lib5c kr are
infile (the countsfile we want to balance) and outfile (where to
write the balanced countsfile).
Processing multiple input files¶
If we want to process a lot of countsfiles, we can use a pattern like this
In [7]:
!lib5c kr 'trimmed/*.counts' kr/%s.counts
lib5c kr trimmed/pNPC_Rep1.counts 'kr/pNPC_Rep1.counts' --max_iterations '3000' --imputation_size '0'
loading counts
kr balancing
writing counts
lib5c kr trimmed/pNPC_Rep2.counts 'kr/pNPC_Rep2.counts' --max_iterations '3000' --imputation_size '0'
loading counts
kr balancing
writing counts
lib5c kr trimmed/v65_Rep1.counts 'kr/v65_Rep1.counts' --max_iterations '3000' --imputation_size '0'
loading counts
kr balancing
writing counts
lib5c kr trimmed/v65_Rep2.counts 'kr/v65_Rep2.counts' --max_iterations '3000' --imputation_size '0'
loading counts
kr balancing
writing counts
Here instead of a single infile, we have put in a glob-expandable
pattern that will match all the countsfiles in the input directory.
We have to quote this pattern to prevent the shell from expanding it. If
we are processing multiple input countsfiles, then we expect multiple
countsfiles as the output. To specify this easily, we include %s in
the outfile, which will be replaced with the replicate name (derived
automatically from the input filenames).
If you’re running this in a cluster environment with the LSF (aka
bsub) job scheduler available, you can install the bsub Python
package via
$ pip install bsub
and then all the input files will be processed in parallel via bsub.
If you aren’t using the LSF job scheduler, you can still specify multiple input files, but they will be processed in series.
Plotting heatmaps¶
Let’s take a look at the countsfiles we just generated.
One way to visualize countsfiles is to plot heatmaps of them. To do this, we can run
In [8]:
!lib5c plot heatmap -p trimmed/primers_trimmed.bed --region Sox2 kr/pNPC_Rep2.counts kr/pNPC_Rep2_Sox2.png
loading counts
preparing to plot
plotting
The resulting image should look something like this
In [9]:
from IPython.display import Image
Image(filename='kr/pNPC_Rep2_Sox2.png', width=400)
Out[9]:
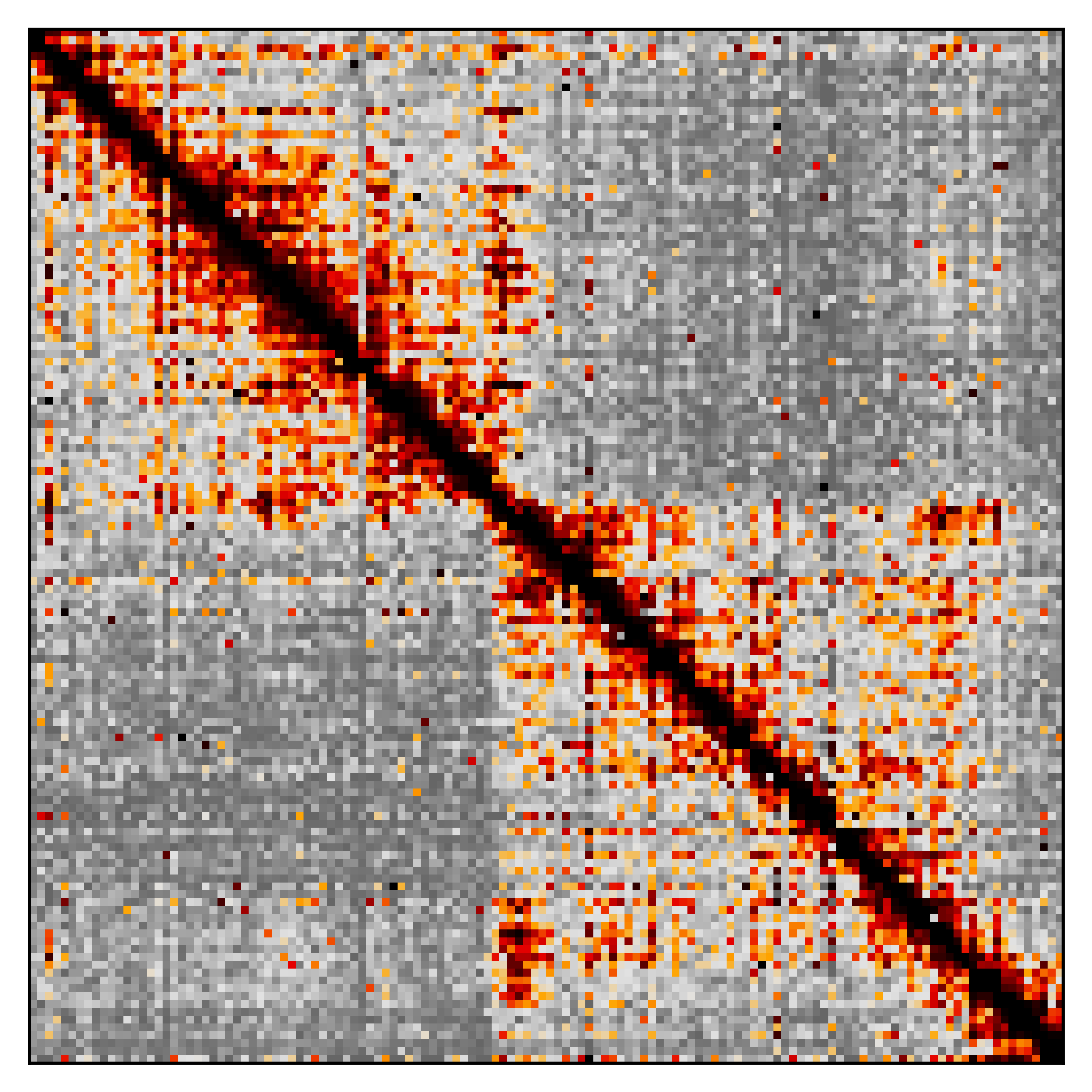
The lib5c plot heatmap subcommand is a lot like the lib5c kr
subcommand. In fact, most lib5c subcommands work in almost exactly
the same way, so learning to use one can help you understand the others.
Specifying a primerfile/binfile with -p/--primerfile¶
Almost every lib5c subcommand that takes countsfiles as its input
also requires a primerfile or a bedfile that it uses to make sure the
countsfiles get parsed correctly. This is accomplished with the -p
flag. We didn’t have to use the -p flag in the lib5c kr example
above because the primerfile we wanted to use was already in the same
directory as the countsfiles, so lib5c was able to find it
automatically. In general though, we might be trying to process
countsfiles that are separated (directory-wise) from the primerfile that
describes their genomic loci, in which case we would have to use the
-p flag as shown above.
Multiple output files (one per region)¶
Some lib5c subcommands generate multiple output files (typically one
per region). To create an output file for only one region, use the
--region (shorter form -r) as shown above. Alternatively, if we
want to generate output for all regions, we can skip the -r/--region
flag and include a %r in the outfile instead, which will be
replaced with the region name. Combining this with the “multiple input
files” idea from above, we can write something like
In [10]:
!lib5c plot heatmap -p trimmed/primers_trimmed.bed 'kr/*.counts' kr/%s_%r.png
lib5c plot heatmap --primerfile 'trimmed/primers_trimmed.bed' --level 'auto' kr/pNPC_Rep1.counts 'kr/pNPC_Rep1_%r.png' --colormap 'obs' --log_base 'None' --pseudocount '1.0'
loading counts
preparing to plot
plotting
lib5c plot heatmap --primerfile 'trimmed/primers_trimmed.bed' --level 'auto' kr/pNPC_Rep2.counts 'kr/pNPC_Rep2_%r.png' --colormap 'obs' --log_base 'None' --pseudocount '1.0'
loading counts
preparing to plot
plotting
lib5c plot heatmap --primerfile 'trimmed/primers_trimmed.bed' --level 'auto' kr/v65_Rep1.counts 'kr/v65_Rep1_%r.png' --colormap 'obs' --log_base 'None' --pseudocount '1.0'
loading counts
preparing to plot
plotting
lib5c plot heatmap --primerfile 'trimmed/primers_trimmed.bed' --level 'auto' kr/v65_Rep2.counts 'kr/v65_Rep2_%r.png' --colormap 'obs' --log_base 'None' --pseudocount '1.0'
loading counts
preparing to plot
plotting
Binning¶
Let’s bin our Knight-Ruiz balanced data!
Before we do this, we need to generate some bins to tile our 5C regions. To do this, we run
In [11]:
!lib5c determine-bins -w 4000 trimmed/primers_trimmed.bed bedfiles/4kb_bins.bed
creating directory bedfiles
The -w/--bin_width flag specifies the width of each bin in base
pairs, and bedfiles/4kb_bins.bed specifies the name of the
newly-created bin bedfile.
Now that we have a bin bedfile describing the bins, we can figure out what the counts values for each bin should be by running
In [12]:
!lib5c bin -w 24000 -p trimmed/primers_trimmed.bed -b bedfiles/4kb_bins.bed kr/pNPC_Rep2.counts binned/pNPC_Rep2.counts
creating directory binned
Notice that the lib5c bin subcommand needs both a primerfile
(specified with -p) and a bin bedfile (specified with -b). It
needs the primerfile to parse the input files, but it also needs a bin
bedfile to know where the bins should be and to write the output files.
The last two arguments are just the input file and the output file, just
like in the previous subcommands.
We can plot a heatmap of our binned data by running
In [13]:
!lib5c plot heatmap -p bedfiles/4kb_bins.bed -r Sox2 -R -g mm9 binned/pNPC_Rep2.counts binned/pNPC_Rep2_Sox2.png
loading counts
preparing to plot
plotting
The result should look something like this
In [14]:
Image(filename='binned/pNPC_Rep2_Sox2.png', width=500)
Out[14]:
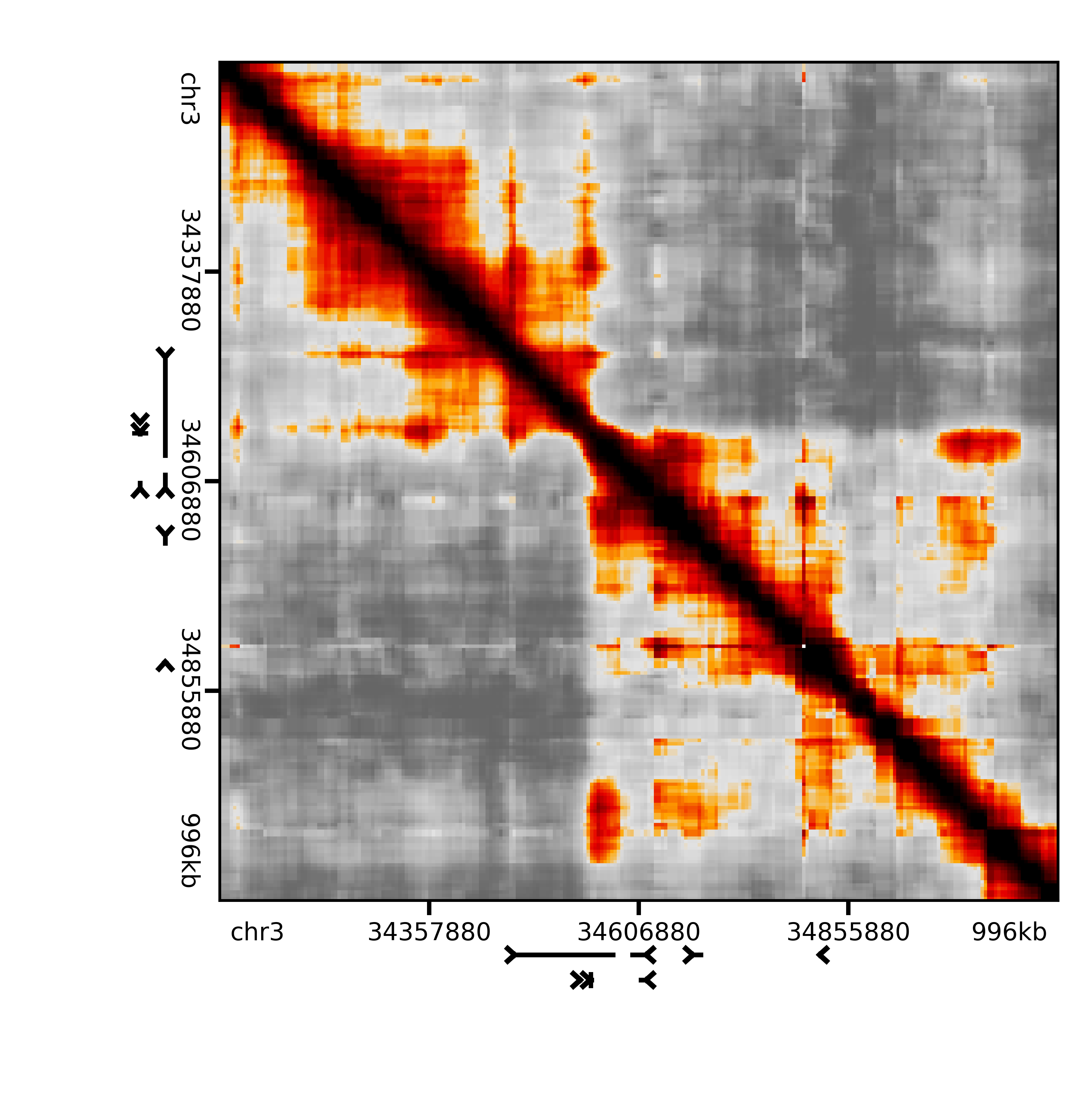
Expected modeling¶
In order to identify “enriched” interactions in our freshly-binned data, we first need to answer the question: “Enriched with respect to what?” The answer to this question is an expected model, which represents our understanding of what we would expect the interaction frequency between any two bins to be. This expected value depends both on the linear genomic separation between the bins, as well as the effects of local contact domain structure.
We can generate an expected model by running
In [15]:
!lib5c expected -p bedfiles/4kb_bins.bed -RED binned/pNPC_Rep2.counts expected/pNPC_Rep2.counts
loading counts
using polynomial log-log 1-D distance model
using polynomial log-log 1-D distance model
applying donut correction
applying donut correction
creating directory expected
Note that for consistency, the flag used to specify the bin bedfile
bedfiles/4kb_bins.bed is still -p. Most lib5c subcommands
work on both fragment-level and bin-level data, with essentially no
changes to the parameters.
We are using the -R/--regression, -E/--exclude_near_diagonal and
-D/--donut_correction flags to indicate that we want to start with a
regional expected model fitted with a simple log-log regression which
ignores points too close to the diagonal, and then apply donut
correction to account for local variations in contact domain structure.
Variance modeling¶
Armed with an expected model, we can now readily identify which pairs of bins interact more frequently than expected. However, what we really want to know is which pairs of bins interact significantly more frequently than expected. In order to understand the extent of insignificant, noise-driven variations at each point, we need to construct a variance model - a statistical variance estimate for each pair of bins.
We can generate a variance model by running
In [16]:
!lib5c variance -p bedfiles/4kb_bins.bed binned/pNPC_Rep2.counts expected/pNPC_Rep2.counts variance/pNPC_Rep2.counts
loading counts
writing variance
creating directory variance
By default, this will fit a log-normal deviation-based distance-variance relationship to the observed data.
P-value calling¶
Finally, we will call p-values for the interactions in our dataset
In [17]:
!lib5c pvalues -p bedfiles/4kb_bins.bed binned/pNPC_Rep2.counts expected/pNPC_Rep2.counts variance/pNPC_Rep2.counts -L norm pvalues/pNPC_Rep2.counts
loading counts
calling pvalues
writing p-values
creating directory pvalues
Calling p-values simply parametrizes a distribution of a specified
family (here we chose -L norm for the lognormal distribution) with
mean value taken from the expected model and variance taken from the
variance model, and then calls a right-tail p-value for the observed
value in the binned data.
P-value heatmaps¶
To visualize p-values, we can use the lib5c plot heatmap subcommand
as above, but we need to invoke it like
In [18]:
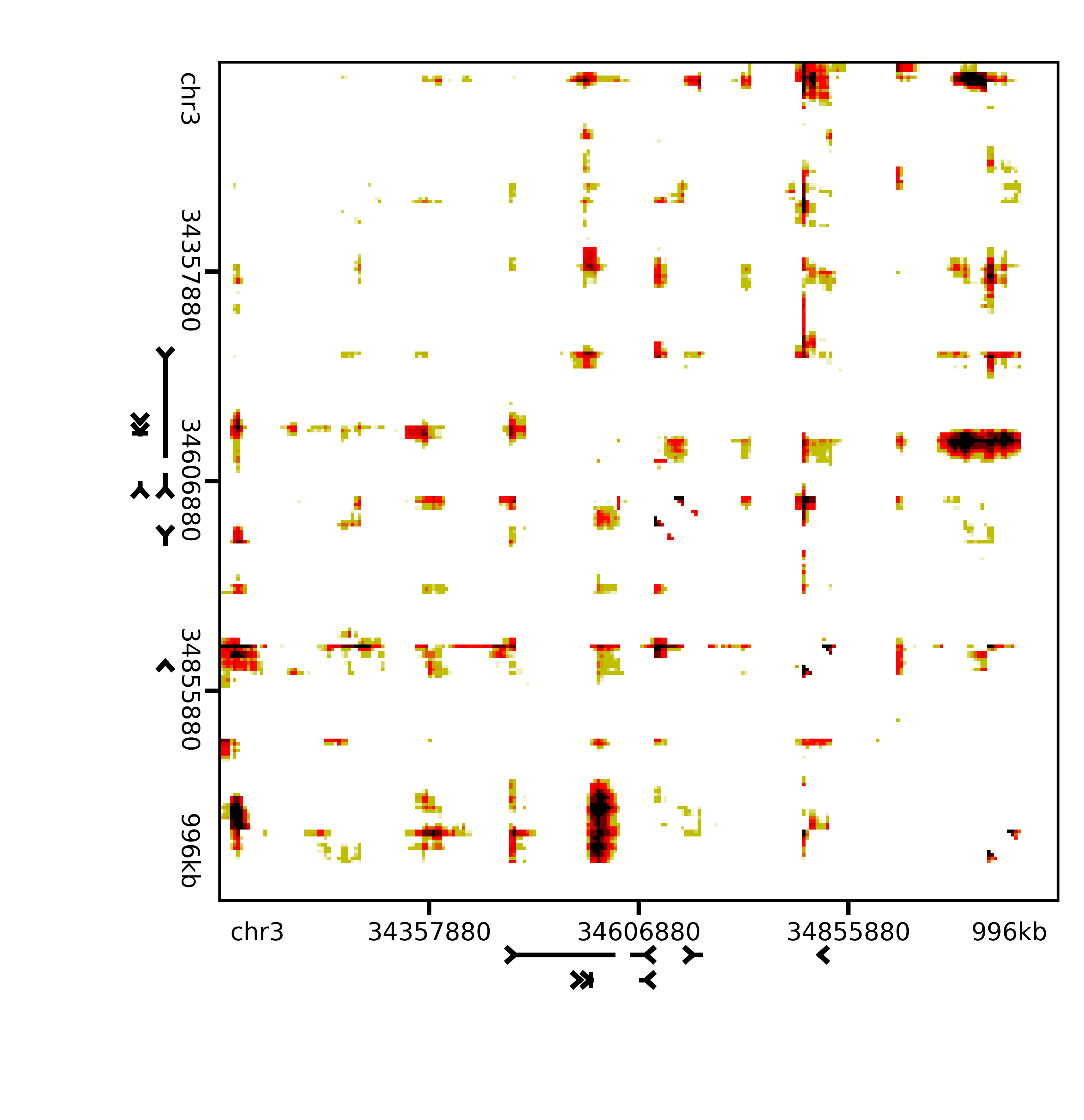
!lib5c plot heatmap -p bedfiles/4kb_bins.bed -r Sox2 -PR -g mm9 pvalues/pNPC_Rep2.counts pvalues/pNPC_Rep2_Sox2.png
loading counts
preparing to plot
plotting
Here we are using the -P/--pvalue flag to indicate that we are
trying to visualize p-values, the -R/--rulers flag to indicate that
we want to add genomic coordinate rulers to the heatmap, and the
-g/--genes flag to indicate that we want to add gene tracks from the
mm9 reference genome.
The results should look something like this:
In [19]:
Image(filename='pvalues/pNPC_Rep2_Sox2.png', width=500)
Out[19]:
Next steps¶
Go ahead and explore the other lib5c subcommands! Remember that you
can list all the subcommands and get detailed help for a particular
subcommand with the -h/--help flag.