Pipeline tutorial¶
This tutorial will walk you through the basic usage of the lib5c
pipeline.
Follow along in Google colab¶
You can run and modify the cells in this notebook tutorial live using Google colaboratory by clicking the link below:
To simply have all the cells run automatically, click
Runtime > Run all in the colab toolbar.
Make sure lib5c is installed¶
Inside a fresh virtual environment, run
In [1]:
!pip install lib5c
Requirement already satisfied: lib5c in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (0.5.3)
Requirement already satisfied: numpy>=1.10.4 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.14.2)
Requirement already satisfied: pandas>=0.18.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.22.0)
Requirement already satisfied: statsmodels>=0.6.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.8.0)
Requirement already satisfied: dill>=0.2.5 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.2.7.1)
Requirement already satisfied: luigi>=2.1.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.7.3)
Requirement already satisfied: matplotlib>=1.4.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.2.2)
Requirement already satisfied: python-daemon<2.2.0,>=2.1.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (2.1.2)
Requirement already satisfied: powerlaw>=1.4.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.4.3)
Requirement already satisfied: interlap>=0.2.3 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.2.6)
Requirement already satisfied: seaborn>=0.8.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.8.1)
Requirement already satisfied: decorator>=4.0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (4.2.1)
Requirement already satisfied: scikit-learn>=0.17.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (0.19.1)
Requirement already satisfied: scipy>=0.16.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from lib5c) (1.0.0)
Requirement already satisfied: python-dateutil in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from pandas>=0.18.0->lib5c) (2.7.2)
Requirement already satisfied: pytz>=2011k in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from pandas>=0.18.0->lib5c) (2018.3)
Requirement already satisfied: patsy in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from statsmodels>=0.6.1->lib5c) (0.5.0)
Requirement already satisfied: tornado<5,>=4.0 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from luigi>=2.1.1->lib5c) (4.5.3)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (2.2.0)
Requirement already satisfied: backports.functools-lru-cache in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.5)
Requirement already satisfied: subprocess32 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (3.5.3)
Requirement already satisfied: six>=1.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.11.0)
Requirement already satisfied: kiwisolver>=1.0.1 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (1.0.1)
Requirement already satisfied: cycler>=0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from matplotlib>=1.4.3->lib5c) (0.10.0)
Requirement already satisfied: setuptools in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (40.4.3)
Requirement already satisfied: docutils in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (0.14)
Requirement already satisfied: lockfile>=0.10 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from python-daemon<2.2.0,>=2.1.1->lib5c) (0.12.2)
Requirement already satisfied: backports-abc>=0.4 in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (0.5)
Requirement already satisfied: certifi in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (2018.1.18)
Requirement already satisfied: singledispatch in /mnt/c/Users/thoma/venv-doc/lib/python2.7/site-packages (from tornado<5,>=4.0->luigi>=2.1.1->lib5c) (3.4.0.3)
Make a directory¶
Create a directory for this pipeline and change into it
$ mkdir lib5c-tutorial
$ cd lib5c-tutorial
Get data¶
First, download some data from GEO. For this tutorial, we will use the 5C data set from GEO Series GSE68582.
The specific files we will need are:
- A primerfile describing the fragments in the 5C assay.
- Four raw countsfiles containing the results of two replicates each of two conditions (v65 and pNPC):
- v65 Rep 1
- v65 Rep 2
- pNPC Rep 1
- pNPC Rep 2
Download the five files using the links above, then unzip them into a
new directory, lib5c-tutorial/input. Finally, strip all of the
.txt extensions and GEO identifiers for brevity. If you use the
bash shell, you can do all of this by running the following
commands:
In [2]:
!mkdir input
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSE68582&format=file&file=GSE68582%5FBED%5FES%2DNPC%2DiPS%2DLOCI%5Fmm9%2Ebed%2Egz' | gunzip -c > input/BED_ES-NPC-iPS-LOCI_mm9.bed
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974095&format=file&file=GSM1974095%5Fv65%5FRep1%2Ecounts%2Etxt%2Egz' | gunzip -c > input/v65_Rep1.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974096&format=file&file=GSM1974096%5Fv65%5FRep2%2Ecounts%2Etxt%2Egz' | gunzip -c > input/v65_Rep2.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974099&format=file&file=GSM1974099%5FpNPC%5FRep1%2Ecounts%2Etxt%2Egz' | gunzip -c > input/pNPC_Rep1.counts
!wget -qO- 'http://www.ncbi.nlm.nih.gov/geo/download/?acc=GSM1974100&format=file&file=GSM1974100%5FpNPC%5FRep2%2Ecounts%2Etxt%2Egz' | gunzip -c > input/pNPC_Rep2.counts
mkdir: cannot create directory ‘input’: File exists
or you can download, unzip, and rename them manually. After renaming,
the lib5c-tutorial/input directory should contain the following
files:
BED_ES-NPC-iPS-LOCI_mm9.bedv65_Rep1.countsv65_Rep2.countspNPC_Rep1.countspNPC_Rep2.counts
Note for Docker image users¶
If you are using lib5c from the Docker image, run
$ docker run -it -v <full path to lib5c-tutorial>:/lib5c-tutorial creminslab/lib5c:latest
root@<container_id>:/# cd /lib5c-tutorial
and continue running all tutorial commands in this shell.
Get the default config file¶
From inside lib5c-tutorial, run lib5c pipeline to drop the
example pipeline configuration file, luigi.cfg.
In [3]:
!lib5c pipeline
no luigi.cfg found in current directory
dropping example luigi.cfg file...
The behavior of the entire pipeline is controlled using this one configuration file.
Edit the config file¶
Open luigi.cfg in your favorite text editor and take a look inside.
The config file is organized into blocks that start with a header and then define some config variables.
The first block is the [PipelineTask] block, which lists the
pipelines to be run in a list called tasks, the entries of which
look like directories. These are in fact the directory structures into
which the outputs will be written. The folder names in the directory
path correspond to individual steps of the pipeline, as described in the
table mapping, which has the general format
"folder_name": ["step_name", {non_default_parameters}]
The same step can be defined with different parameters, as long as the output folder names are distinguishable; for example, the section
...
"bin_gmean_16_4": ["MakeBinned", {}],
"bin_amean_20_8": ["MakeBinned", {"window_function": "amean",
"bin_width": 8000,
"window_width": 20000}],
...
shows two different parameter choices for the binning step.
The second block is the [RawCounts] block, which specifies where the
input countsfiles are located on the disk and gives them replicate
names.
The third block is the [PrimerFile] block, which specifies where to
find the primerfile on the disk.
The remaining sections expose the default parameters for each step
(e.g., [MakeBinned] describes the default binning step parameters).
For this tutorial, you can leave the [PipelineTask] block unchanged.
However, we need to fill in the [RawCounts] and [PrimerFile]
blocks with the paths to the data files we downloaded. For example:
[RawCounts]
countsfiles={
"ES_Rep1": "input/v65_Rep1.counts",
"ES_Rep2": "input/v65_Rep2.counts",
"pNPC_Rep1": "input/pNPC_Rep1.counts",
"pNPC_Rep2": "input/pNPC_Rep2.counts"
}
[PrimerFile]
primerfile=input/BED_ES-NPC-iPS-LOCI_mm9.bed
The config parser is somewhat sensitive to indentation, so match this carefully.
The replicate names are the keys of the countsfiles dictionary. You
can set them to anything you like; however, the “conditions” (for the
purposes of classifying condition-specific interactions) as defined in
the conditions comma-separated list under the [MakeThreshold]
block near the bottom of the config file must be substrings of the
replicate names. For convenience, by default conditions=ES,pNPC
which matches the replicate names we have chosen above, but when you do
your own analyses you may need to change this.
In [4]:
!sed -i 's/"rep1": "path\/to\/rep1.counts",/"ES_Rep1": "input\/v65_Rep1.counts",\n "ES_Rep2": "input\/v65_Rep2.counts",/g' luigi.cfg
!sed -i 's/"rep2": "path\/to\/rep2.counts"/"pNPC_Rep1": "input\/pNPC_Rep1.counts",\n "pNPC_Rep2": "input\/pNPC_Rep2.counts"/g' luigi.cfg
!sed -i 's/path\/to\/primerfile.bed/input\/BED_ES-NPC-iPS-LOCI_mm9.bed/g' luigi.cfg
Just to make the notebook run faster, we’ll disable some of the automatic plot generation and focus on just two regions
In [5]:
!sed -i '/plot_outfile=%d\/expected-plots\/%s_%r.png/d' luigi.cfg
!sed -i 'N;s/outfile_pattern=%s_qnorm.counts\nheatmap=True/outfile_pattern=%s_qnorm.counts\nheatmap=False/g' luigi.cfg
!sed -i 'N;s/outfile_pattern=%s_express.counts\nheatmap=True/outfile_pattern=%s_express.counts\nheatmap=False/g' luigi.cfg
!sed -i '/Sox2\|Klf4/!d' input/BED_ES-NPC-iPS-LOCI_mm9.bed
After specifying the input files, save the config file, then run
In [6]:
!lib5c pipeline
creating directory ./raw
lib5c plot heatmap -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -R ./raw/ES_Rep1_raw.counts ./raw/ES_Rep1_raw_%r.png
loading counts
preparing to plot
plotting
lib5c plot heatmap -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -R ./raw/pNPC_Rep2_raw.counts ./raw/pNPC_Rep2_raw_%r.png
loading counts
preparing to plot
plotting
lib5c plot heatmap -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -R ./raw/ES_Rep2_raw.counts ./raw/ES_Rep2_raw_%r.png
loading counts
preparing to plot
plotting
lib5c plot heatmap -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -R ./raw/pNPC_Rep1_raw.counts ./raw/pNPC_Rep1_raw_%r.png
loading counts
preparing to plot
plotting
lib5c qnorm -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -R ./raw/qnormed/%s_qnorm.counts ./raw/pNPC_Rep2_raw.counts ./raw/ES_Rep2_raw.counts ./raw/ES_Rep1_raw.counts ./raw/pNPC_Rep1_raw.counts
loading counts
quantile normalizing
writing counts
lib5c express -J -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -B './raw/qnormed/*_qnorm.counts' ./raw/qnormed/jointexpress/%s_express.counts
loading counts
precomputing expected models
joint express normalizing
writing counts
writing bias vectors
lib5c bin -b bedfiles/4kb_bins.bed -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -f gmean -w 16000 -t 0.0 -W ./raw/qnormed/jointexpress/pNPC_Rep1_express.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned.counts
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c expected -p bedfiles/4kb_bins.bed -d 1 -f 0.8 -w 15 -x 5 -m 0.2 -e 0.1 -t auto -M -E -D -O -X ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep1_expected.counts
loading counts
defaulting to logged empirical binned 1-D distance model
defaulting to logged empirical binned 1-D distance model
forcing monotonicity
forcing monotonicity
applying donut correction
applying donut correction
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep1_expected_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c variance -p bedfiles/4kb_bins.bed -m lognorm -s deviation -f lowess -a lowess --min_obs 2.0 --min_disp 1e-8 --min_dist 6 --x_unit dist --y_unit disp ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pNPC_Rep1_variance.counts
loading counts
writing variance
lib5c pvalues -p bedfiles/4kb_bins.bed ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pNPC_Rep1_variance.counts nbinom ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep1_pvalues.counts
loading counts
calling pvalues
writing p-values
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep1_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep1_pvalues_%r.png -P
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c bin -b bedfiles/4kb_bins.bed -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -f gmean -w 16000 -t 0.0 -W ./raw/qnormed/jointexpress/ES_Rep1_express.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned.counts
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c expected -p bedfiles/4kb_bins.bed -d 1 -f 0.8 -w 15 -x 5 -m 0.2 -e 0.1 -t auto -M -E -D -O -X ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep1_expected.counts
loading counts
defaulting to logged empirical binned 1-D distance model
defaulting to logged empirical binned 1-D distance model
forcing monotonicity
forcing monotonicity
applying donut correction
applying donut correction
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep1_expected_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c variance -p bedfiles/4kb_bins.bed -m lognorm -s deviation -f lowess -a lowess --min_obs 2.0 --min_disp 1e-8 --min_dist 6 --x_unit dist --y_unit disp ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/ES_Rep1_variance.counts
loading counts
writing variance
lib5c pvalues -p bedfiles/4kb_bins.bed ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep1_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep1_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/ES_Rep1_variance.counts nbinom ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep1_pvalues.counts
loading counts
calling pvalues
writing p-values
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep1_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep1_pvalues_%r.png -P
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c bin -b bedfiles/4kb_bins.bed -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -f gmean -w 16000 -t 0.0 -W ./raw/qnormed/jointexpress/ES_Rep2_express.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned.counts
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c expected -p bedfiles/4kb_bins.bed -d 1 -f 0.8 -w 15 -x 5 -m 0.2 -e 0.1 -t auto -M -E -D -O -X ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep2_expected.counts
loading counts
defaulting to logged empirical binned 1-D distance model
defaulting to logged empirical binned 1-D distance model
forcing monotonicity
forcing monotonicity
applying donut correction
applying donut correction
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep2_expected_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c variance -p bedfiles/4kb_bins.bed -m lognorm -s deviation -f lowess -a lowess --min_obs 2.0 --min_disp 1e-8 --min_dist 6 --x_unit dist --y_unit disp ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/ES_Rep2_variance.counts
loading counts
writing variance
lib5c pvalues -p bedfiles/4kb_bins.bed ./raw/qnormed/jointexpress/bin_gmean_16_4/ES_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/ES_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/ES_Rep2_variance.counts nbinom ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep2_pvalues.counts
loading counts
calling pvalues
writing p-values
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep2_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep2_pvalues_%r.png -P
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c bin -b bedfiles/4kb_bins.bed -p input/BED_ES-NPC-iPS-LOCI_mm9.bed -f gmean -w 16000 -t 0.0 -W ./raw/qnormed/jointexpress/pNPC_Rep2_express.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned.counts
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c expected -p bedfiles/4kb_bins.bed -d 1 -f 0.8 -w 15 -x 5 -m 0.2 -e 0.1 -t auto -M -E -D -O -X ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep2_expected.counts
loading counts
defaulting to logged empirical binned 1-D distance model
defaulting to logged empirical binned 1-D distance model
forcing monotonicity
forcing monotonicity
applying donut correction
applying donut correction
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep2_expected_%r.png
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c variance -p bedfiles/4kb_bins.bed -m lognorm -s deviation -f lowess -a lowess --min_obs 2.0 --min_disp 1e-8 --min_dist 6 --x_unit dist --y_unit disp ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pNPC_Rep2_variance.counts
loading counts
writing variance
lib5c pvalues -p bedfiles/4kb_bins.bed ./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/pNPC_Rep2_expected.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pNPC_Rep2_variance.counts nbinom ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep2_pvalues.counts
loading counts
calling pvalues
writing p-values
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep2_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep2_pvalues_%r.png -P
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
lib5c threshold -p bedfiles/4kb_bins.bed -c 'ES,pNPC' -t 1e-15 -s 3 -d 24000 -b 0.6 -o ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/dataset.tsv -k ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/kappa_confusion.txt ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/classifications.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep2_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep2_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/ES_Rep1_pvalues.counts ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep1_pvalues.counts
loading counts
thresholding
preparing dataset
assigning rep ES_Rep1 to class ES
assigning rep ES_Rep2 to class ES
assigning rep pNPC_Rep1 to class pNPC
assigning rep pNPC_Rep2 to class pNPC
distance thresholding
significance thresholding
size thresholding
intersecting across conditions
adding background
counting final clusters
writing results
lib5c plot heatmap -p bedfiles/4kb_bins.bed -R './raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/classifications.counts' ./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/classifications_%r.png -TC
loading counts
preparing to plot
plotting
encountered exception, falling back to series operation
a second time to run the pipeline.
Once the pipeline completes, output files should be present in subfolders of the directory you ran the pipeline in. Look around and investigate the results of your analysis!
The pipeline output is written into a tree of folders representing the stages of the pipeline. Each folder stores the data files for all replicates at that stage, as well as some optional heatmap visualizations for each replicate and region. We’ll check out a few of them below.
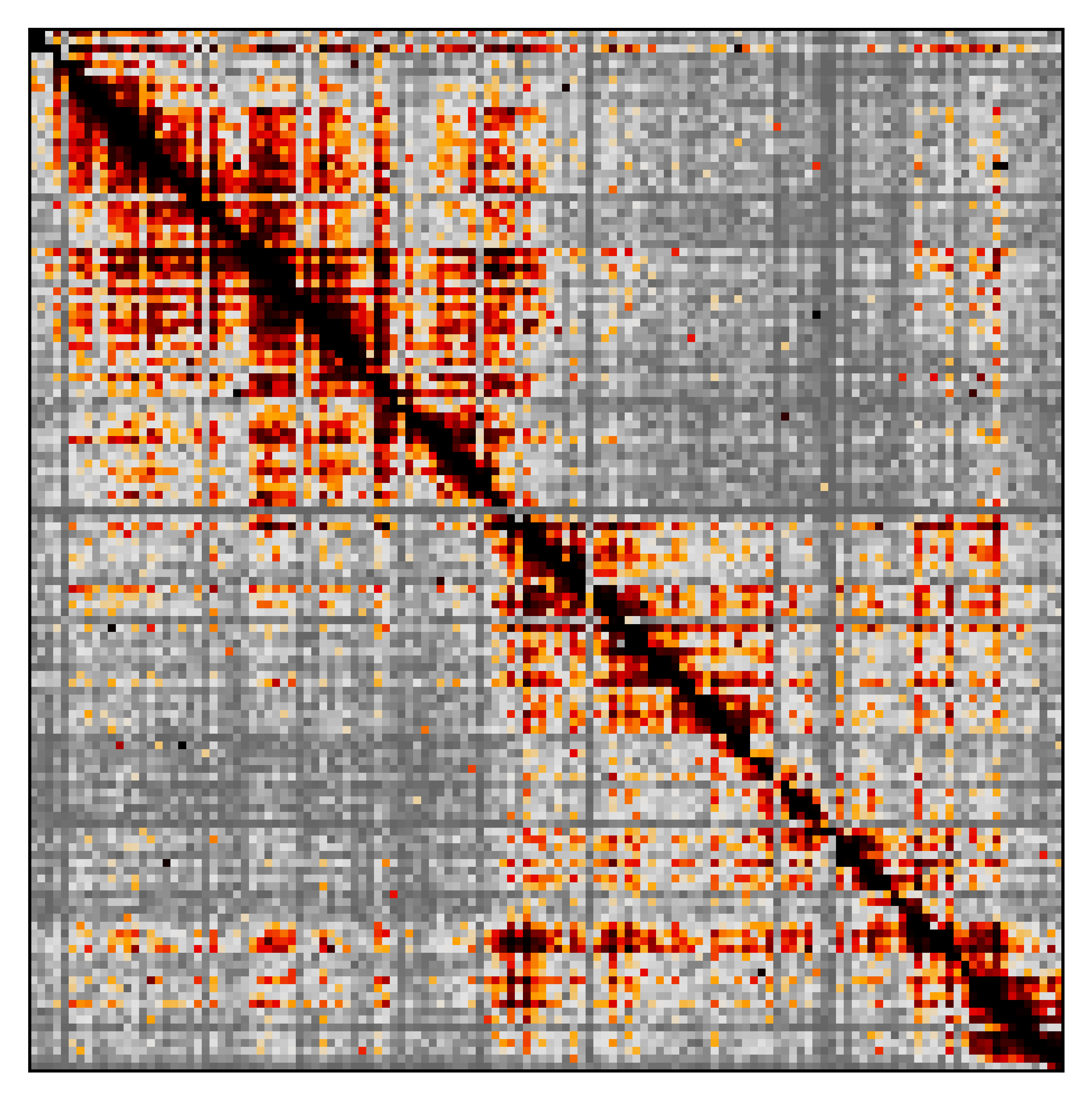
Here’s the raw heatmap for the Sox2 region for pNPC2 Rep2:
In [7]:
from IPython.display import Image
Image(filename='./raw/pNPC_Rep2_raw_Sox2.png', width=400)
Out[7]:
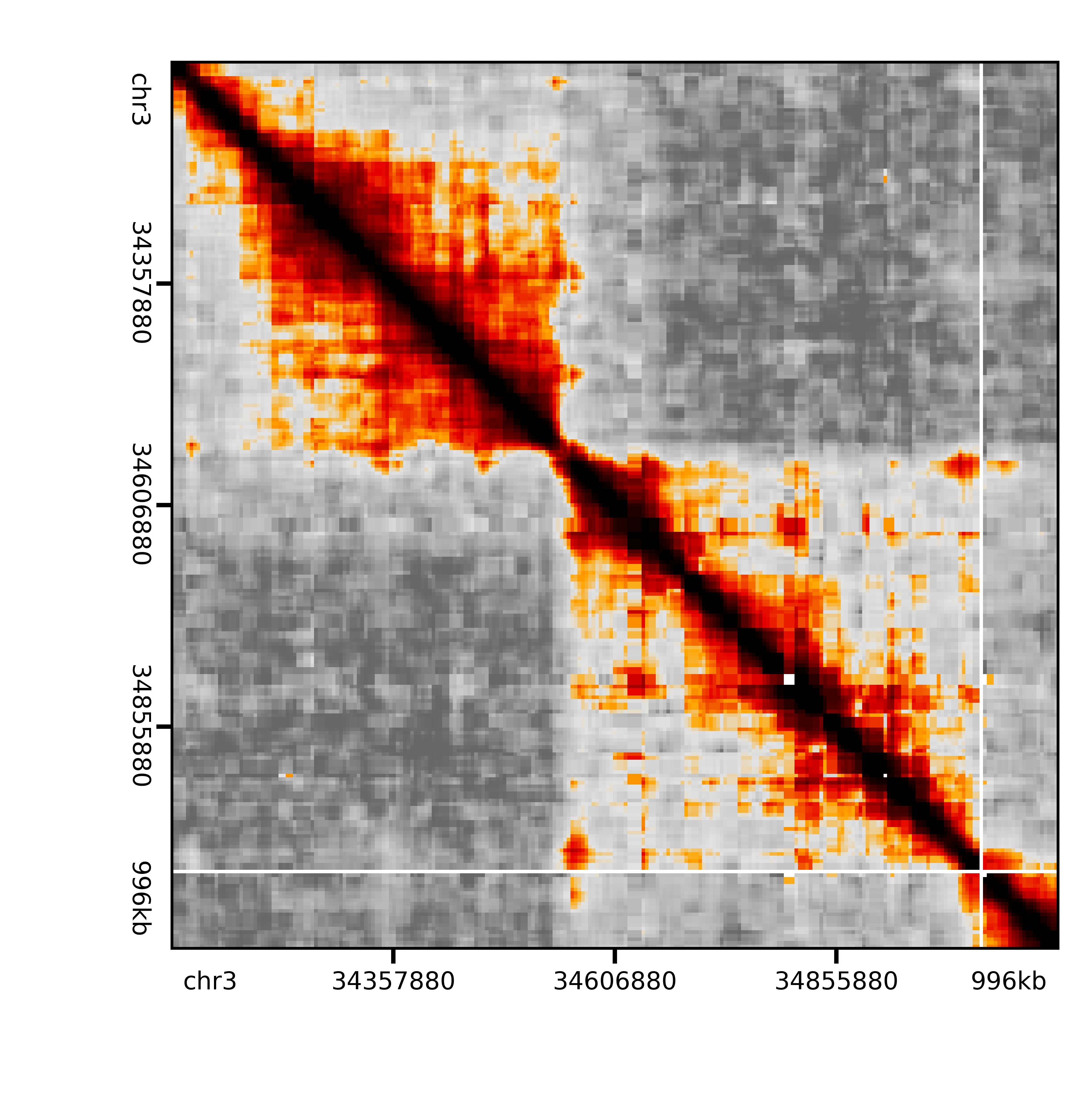
Here’s the same rep and region after balancing and binning:
In [8]:
Image(filename='./raw/qnormed/jointexpress/bin_gmean_16_4/pNPC_Rep2_binned_Sox2.png', width=400)
Out[8]:
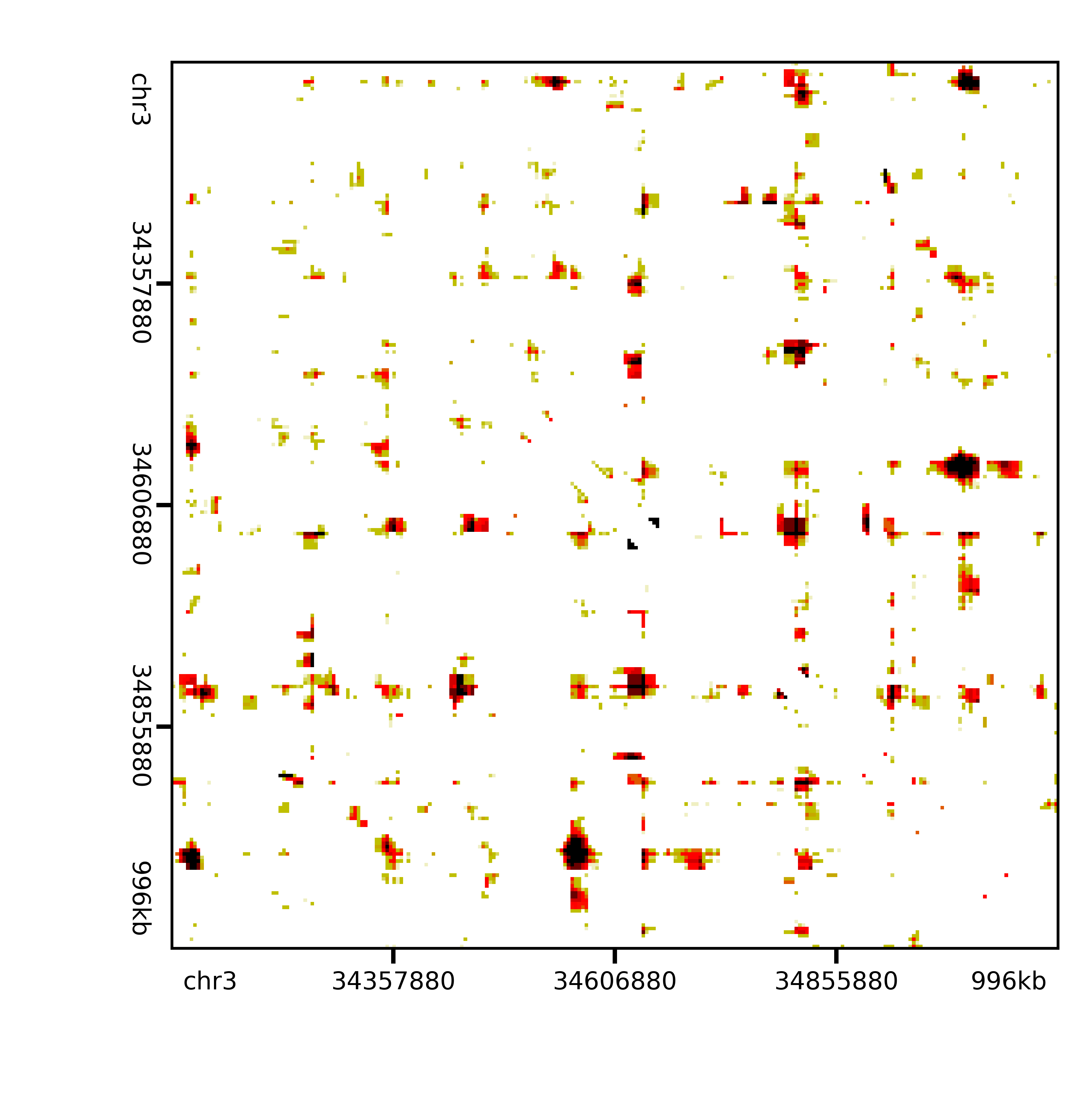
and after expected and variance modeling and p-value calling:
In [9]:
Image(filename='./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/pNPC_Rep2_pvalues_Sox2.png', width=400)
Out[9]:
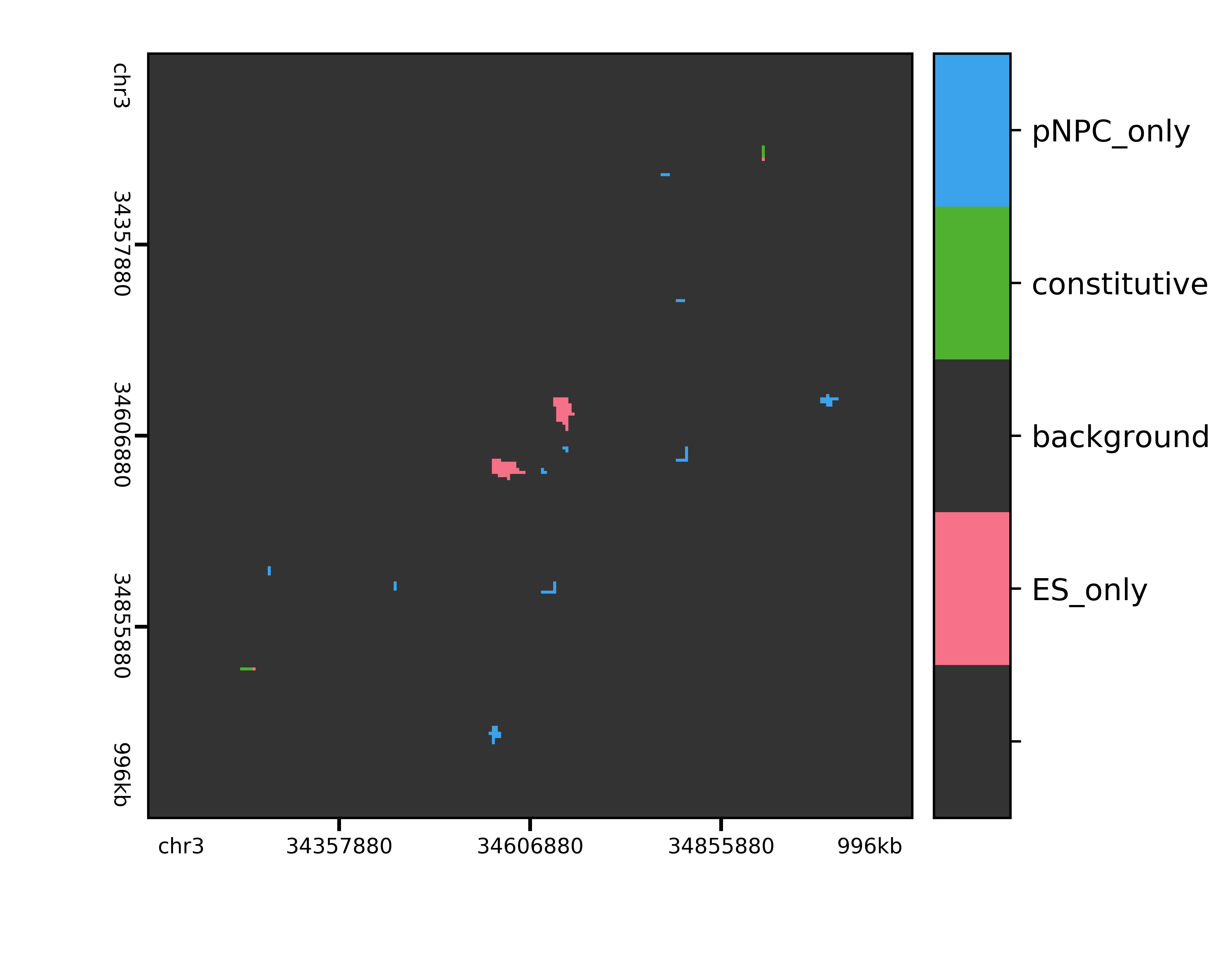
Finally, here are the final condition-specific cluster calls:
In [10]:
Image(filename='./raw/qnormed/jointexpress/bin_gmean_16_4/expected_donut/variance/pvalues/threshold/classifications_Sox2.png', width=600)
Out[10]: